
Catalog excerpts

Bioinformatics Toolbox Read, analyze, and visualize genomic and proteomic data Bioinformatics Toolbox™ provides algorithms and visualization techniques for Next Generation Sequencing (NGS), microarray analysis, mass spectrometry, and gene ontology. Using toolbox functions, you can read genomic and proteomic data from standard file formats such as SAM, FASTA, CEL, and CDF, as well as from online databases such as the NCBI Gene Expression Omnibus and GenBank®. You can explore and visualize this data with sequence browsers, spatial heatmaps, and clustergrams. The toolbox also provides statistical techniques for detecting peaks, imputing values for missing data, and selecting features. You can combine toolbox functions to support common bioinformatics workflows. You can use ChIP-Seq data to identify transcription factors; analyze RNA-Seq data to identify differentially expressed genes; identify copy number variants and SNPs in microarray data; and classify protein profiles using mass spectrometry data. Learn more about computational biology. Key Features ▪ Next Generation Sequencing analysis and browser ▪ Sequence analysis and visualization, including pairwise and multiple sequence alignment and peak detection ▪ Microarray data analysis, including reading, filtering, normalizing, and visualization ▪ Mass spectrometry analysis, including preprocessing, classification, and marker identification ▪ Phylogenetic tree analysis ▪ Graph theory functions, including interaction maps, hierarchy plots, and pathways ▪ Data import from genomic, proteomic, and gene expression files, including SAM, FASTA, CEL, and CDF, and from databases such as NCBI and GenBank What additional features would you like in Bioinformatics Toolbox?
Open the catalog to page 1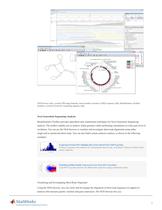
NGS browser (top), circular DNA map (bottom), and secondary structure of RNA sequence (left). Bioinformatics Toolbox includes a variety of tools for visualizing sequence data. Next Generation Sequencing Analysis Bioinformatics Toolbox provides algorithms and visualization techniques for Next Generation Sequencing analysis. The toolbox enables you to analyze whole genomes while performing calculations at a base pair level of resolution. You can use the NGS browser to visualize and investigate short-read alignments using either single-end or paired-end short reads. You can also build custom...
Open the catalog to page 2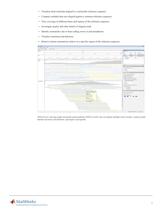
■ Visualize short-read data aligned to a nucleotide reference sequence ■ Compare multiple data sets aligned against a common reference sequence ■ View coverage of different bases and regions of the reference sequence ■ Investigate quality and other details of aligned reads ■ Identify mismatches due to base-calling errors or polymorphisms ■ Visualize insertions and deletions ■ Retrieve feature annotations relative to a specific region of the reference sequence NGS browser, showing single nucleotide polymorphisms (SNPs) in bold. You can display multiple tracks of data, examine peaks, identify...
Open the catalog to page 3
Custom plot mapping E-box motifs to peaks in a wavelet denoised signal. Storing and Managing Short-Read Sequence Data The data sets used in Next Generation Sequencing analysis are often too large to fit into physical memory. Bioinformatics Toolbox provides specialized data containers that enable you to analyze entire genomes. The BioIndexedFile object lets you access the contents of text files containing nonuniform-sized entries such as sequences, annotations, and cross references to the data set. You can generate these objects from tables, flat files, or application-specific formats such...
Open the catalog to page 4
Data Analysis and Visualization Bioinformatics Toolbox lets you perform background adjustments and calculate gene (probe set) expression values from Affymetrix microarray probe-level data using Robust Multi-Array Average (RMA) and GC Robust Multi-Array Average (GCRMA) procedures. You can apply circular binary segmentation to array CGH data and estimate the false discovery rate of multiple hypotheses testing of gene expression data from a microarray experiment. You can also perform rank-invariant set normalization on either probe intensities for multiple Affymetrix CEL files or gene...
Open the catalog to page 5
BJ Copy Number Variation File b d it View Iniert Tools Of: Hep Window Help Q Human Karyogram with Copy Number AbreraborK Fh Edi1 V"«w Inwrt Iwli Otsktsp Human Karyngram wilh Copy Number Alterations of GMQ5G79 Copy number alterations (left) calculated and viewed alongside ideograms (right) using Bioinformatics Toolbox. Mass Spectrometry Data Analysis Bioinformatics Toolbox provides a set of functions for mass spectrometry data analysis. These functions enable preprocessing, classification, and marker identification from SELDI, MALDI, LC/MS, and GC/MS data. Preprocessing functions include...
Open the catalog to page 6
Label-free differential proteomics and metabolomics analysis using Bioinformatics Toolbox. You can smooth, align, and normalize spectra and then use classification and statistical learning tools to create classifiers and identify potential biomarkers. '* Identifying Significant Features and Classifying Protein Profiles * See classification of mass spectrometry data and statistical tools that can be used to look for ll, 1, ..., I potential disease markers and proteomic pattern diagnostics. Graph Theory, Statistical Learning, and Gene Ontology Graph Theory and Visualization Bioinformatics...
Open the catalog to page 7
Gene Ontology Bioinformatics Toolbox enables you to access the Gene Ontology database from within MATLAB®, parse gene ontology annotated files, and obtain subsets of the ontology such as ancestors, descendants, or relatives. Sequence Analysis Bioinformatics Toolbox provides sequence analysis and visualization tools for genomic and proteomic sequence data. You can perform a variety of analyses, including multiple sequence alignments and the building and interactive viewing and manipulation of phylogenetic trees. Sequence Alignment The toolbox provides functions, objects, and methods for...
Open the catalog to page 8
Protein Feature Analysis The toolbox provides protein sequence analysis techniques, including routines for calculating properties of a peptide sequence such as atomic composition, isoelectric point, and molecular weight. You can determine the amino acid composition of protein sequences, cleave a protein with an enzyme, and create backbone plots and Ramachandran plots of PDB data. You can use the Sequence Tool to view the properties of an amino acid sequence or use the Molecule Viewer to display and manipulate 3D molecular structures. Data Import and Application Deployment File Formats and...
Open the catalog to page 9All The MathWorks catalogs and technical brochures
-
MATLAB Production Server
6 Pages
-
Database Toolbox
4 Pages
-
MATLAB Report Generator
4 Pages
-
Stateflow
8 Pages
-
SimEvents
7 Pages
-
SimDriveline
7 Pages
-
SimHydraulics
7 Pages
-
SimPowerSystems
8 Pages
-
Simulink Control Design
5 Pages
-
Aerospace Blockset
5 Pages
-
SimRF
6 Pages
-
Simulink Coder
6 Pages
-
Embedded Coder
8 Pages
-
Simulink PLC Coder
4 Pages
-
Fixed-Point Designer
9 Pages
-
MATLAB Coder
5 Pages
-
Simulink 3D Animation
10 Pages
-
Gauges Blockset
2 Pages
-
Simulink Report Generator
3 Pages
-
Polyspace Bug Finder
6 Pages
-
global-optimization-toolbox
10 Pages
-
Phased Array System Toolbox
9 Pages
-
OPC Toolbox
5 Pages
-
Simulink Design Verifier
7 Pages
-
Simulink Design Optimization
10 Pages
-
Filter Design HDL Coder
5 Pages
-
SimBiology
6 Pages
-
Computer Vision System Toolbox
10 Pages
-
DSP System Toolbox
11 Pages
-
Fuzzy Logic Toolbox
5 Pages
-
Polyspace Client for C/C++
5 Pages
-
xPC Target
5 Pages
-
SimMechanics
7 Pages
-
Simscape
7 Pages
-
Simulink
6 Pages
-
Data Acquisition Toolbox
8 Pages
-
Image Processing Toolbox
7 Pages
-
Signal Processing Toolbox
10 Pages
-
Control System Toolbox
6 Pages
-
Symbolic Math Toolbox?
6 Pages
-
Parallel Computing Toolbox?
7 Pages
-
MATLAB®
6 Pages
-
Mapping Toolbox 3.2
7 Pages
-
Instrument Control Toolbox
7 Pages
-
Optimization Toolbox 6.0
14 Pages
Archived catalogs
-
MATLAB Release Notes
505 Pages
-
C and Fortran API Reference
263 Pages
-
External Interfaces
649 Pages
-
Function Reference: Volume 3 (P-Z)
1696 Pages
-
Function Reference: Volume 2 (F-O)
1568 Pages
-
Function Reference: Volume 1 (A-E)
1298 Pages
-
Creating Graphical User Interfaces
520 Pages
-
3-D Visualization
212 Pages
-
Graphics
667 Pages
-
MATLAB Programming Tips
66 Pages
-
Programming Fundamentals
840 Pages
-
Data Analysis
220 Pages
-
Mathematics
316 Pages
-
MATLAB® Getting Started Guide
250 Pages





































































