
3D dual-color PALM/dSTORM imaging of centrosomal proteins with nanometric resolution using MicAO 3DSR - Adaptive optics for microscopy Application Notes
1 /
6Pages
Catalog excerpts

3D dual-color PALM/dSTORM imaging of centrosomal proteins with nanometric resolution using MicAO 3DSR Grégory CLOUVEL, Audrius JASAITIS and Xavier LEVECQ Imagine Optic, 18 rue Charles de Gaulle, 91400 Orsay, France contact@imagine-optic.com Summary Photoactivation localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM) are becoming routine methods in biological imaging with optical resolution beyond the light diffraction barrier. We present MicAO 3DSR – the first adaptive optics device which introduces the three dimensional imaging capability for PALM/STORM. This apparatus, designed by Imagine Optic, contains the wave-front sensor and deformable mirror. With the help of these components MicAO 3D-SR corrects various types of aberrations, induced by optical elements inside the microscope and by the biological sample. MicAO 3D-SR optimizes the Point Spread Function (PSF) of the microscope and consequently improves the lateral localization precision of the PALM/STORM setup by 40%. In addition to that, MicAO 3D-SR brings in the ability to image fluorescent molecules in 3D. With its deformable mirror, it introduces controlled perfect astigmatism and allows us to precisely locate the position of fluorescing molecule in all three dimensions. In this application note we demonstrate the localization precision of PALM/STORM method when it is operated together with MicAO 3D-SR by 3D dual-color imaging of two proteins in centrosome – the cellular organelle which dimensions lye beyond the diffraction limit of 250x250x500 nm. 3D dual-color PALM/dSTORM imaging of centrosomal proteins with nanometric resolution using MicAO 3DSR Application note imagine-optic.com 8 December 2015 – Property of Imagine Optic
Open the catalog to page 1
The nanometric resolution of intracellular components has become a necessary requirement in the field of fluorescence microscopy. The question of co-localization of proteins within cellular structures is very important in solving important biological problems. For example visualizing the structure of various adhesion complexes, the structure of nuclear pores, or the localization of different proteins in the cell membrane would help researchers to understand functional mechanisms. Unfortunately, the size of such structures usually lies beyond the diffraction limit and therefore they cannot...
Open the catalog to page 2
Figure 2. Image (averaged over 40 individual beads) of the PSF with 60 nm RMS astigmatism at -250 nm of the focus (left image), at the focus (center image), and at +250 nm of the focus (right image). Centrosome plays an important role in many different cellular processes including the organization of microtubule arrays, cell cycle progression, the establishment of polarity of the cell and formation of primary cilium. One centrosome consists of two centrioles, which are barrel-shaped structures made of microtubules that are approximately 200 nm in diameter and 400–500 nm in length (Robbins...
Open the catalog to page 3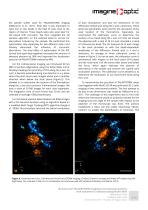
the specific buffer used for PALM/dSTORM imaging (Sillibourne et al , 2011). Note that it was important to have one or two beads in the field of view close to the region of interest. These beads were also used later for the lateral drift correction. We then reapplied the 3N iterative algorithm on the selected bead to correct for aberrations induced by the sample. We performed this optimization step separately for two different colors and thereby eliminated the influence of chromatic aberrations. The total effect of optimization of the PSF (closed and open-loop together) increased the amount...
Open the catalog to page 4
Cep164 component of distal appendages was also chosen as a candidate for simultaneous dSTORM imaging because distal appendages of the mother centrioles are only a few nanometers in width and are challenging to image by immuno-EM. The HeLa mEos2-centrin1 cell line was labeled with an antibody to Cep164, stained with Alexa 647-conjugated secondary antibody and dual color two dimensional PALM/dSTORM imaging was performed (Sillibourne et al, 2011). In this application note, we present the dual-color 3-dimensional imaging of the same centriolar proteins using MicAO 3D-SR. Two different...
Open the catalog to page 5
Sillibourne JE, Specht CG, Izeddin I, Hurbain I, Tran P, Triller A, Darzacq X, Dahan M, Bornens M. (2011) Assessing the Localization of Centrosomal Proteins by PALM/STORM Nanoscopy. Cytoskeleton 68, 619-627. Vorobjev IA, Chentsov Yu S. (1982) Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol, 93, 938–949. Quirin S, Pavani SRP, Piestun R. (2012) Optimal 3D singlemolecule localization for super resolution microscopy with aberrations and engineered point spread functions. PNAS, 109, 675-679. 3D dual-color PALM/dSTORM imaging of centrosomal proteins with nanometric...
Open the catalog to page 6All Imagine Optic catalogs and technical brochures
-
WAVE Suite
3 Pages
Archived catalogs
-
Microtraps
4 Pages
-
AO inside laser chain
5 Pages
-
AO in femtosecond laser
5 Pages
-
Large deformable mirror ILAO
6 Pages
-
NIR optics characterization
6 Pages
-
Telescope characterization
3 Pages
-
absolute measurement
4 Pages
-
HASO R.FLEX
4 Pages
-
HASO3
2 Pages
-
bendAO?
3 Pages
-
HASO?3 WSR Wavefront Sensors
2 Pages
-
HASO R-Flex
3 Pages
-
SL-Sys LIQUID
2 Pages
-
SL-Sys neo
2 Pages
-
HASO™3 Wavefront Sensors
3 Pages































































